Research Projects
As a researcher working at the intersection of artificial intelligence (AI) and biomedical imaging, I focus on both diagnostic applications and the investigation of disease mechanisms, particularly in cancer and neurodegenerative diseases (NDs). My work leverages trustworthy AI to enhance traditional and novel imaging modalities, uncover hidden pathological patterns, and integrate multimodal data. These efforts support drug discovery, improve diagnostic precision, and inform personalized treatment decisions.
AI-driven Understanding of the Brain
I established computational frameworks that support large-scale, quantitative analysis of neurodegenerative disease pathology, open opportunities to identify novel tissue-level markers of disease progression, and broaden exploration in systems neuroscience.
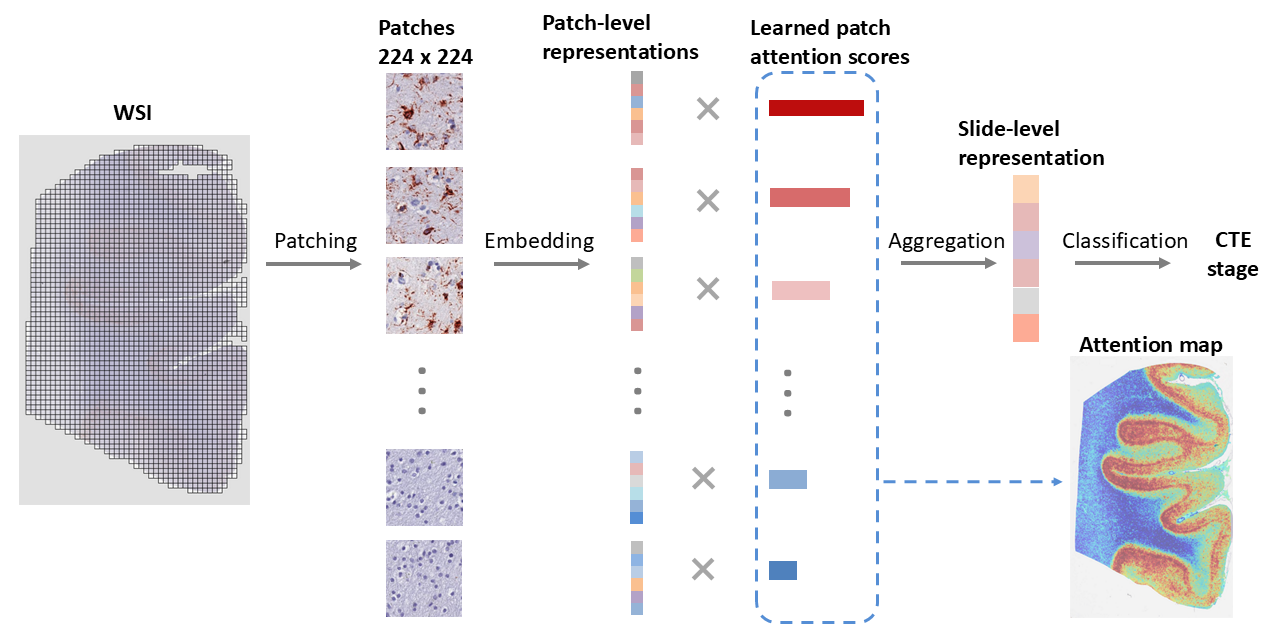
Attention-based weakly-supervised AI for understanding neurodegenerative diseases
I developed data-efficient deep learning models trained with only slide-level supervision to extract biologically meaningful features from neuropathology images. The model generated attention maps that highlighted structural changes linked to tau aggregation, revealing subtle patterns often missed by manual review. These tools support large-scale, quantitative analysis of ND pathology and open new opportunities for identifying novel tissue-level markers of disease progression. [Paper]
A vision foundation model for neuropathology for advancing AI-driven neuropathological image analysis
I am developing the first neuropathology-specific foundation model leveraging the world’s largest digital neuropathology database, aiming to fully unlock the potential of AI-based analysis in advancing our understanding of NDs.

Physics-Informed Neural Network for Mapping Tissue Dynamics Using Laser Speckle Contrast Imaging
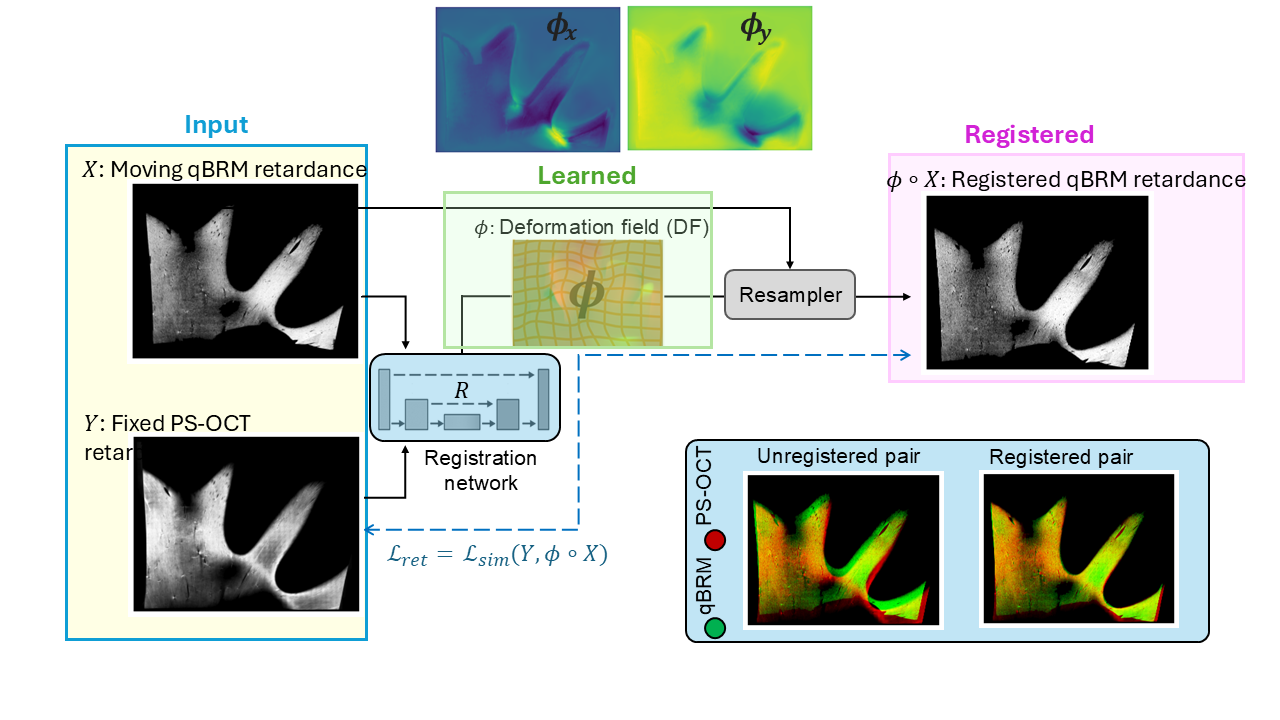
Self-supervised registration of Polarization-Sensitive Optical Coherence Tomography (PS-OCT) and quantitative Birefringence Microscopy (qBRM) images
I developed a self-supervised registration framework to align PS-OCT and qBRM images without requiring paired ground truth. This approach leverages intrinsic structural consistency across modalities to learn accurate spatial correspondence in a data-efficient manner.
Multimodal AI for Breast Cancer Diagnosis
I have developed data fusion approaches to combine ultrasound, Diffuse Optical Tomography (DOT), and radiologists’ BIRADS assessments for breast cancer diagnosis. The proposed two-stage approach can potentially distinguish breast cancers from benign lesions in near real-time. [Paper1: Two-stage] [code1] [Paper2: DOT + BIRADS] [code2] [Paper3: DOT + ultrasound] [code3]


AI for Colorectal Cancer Diagnosis
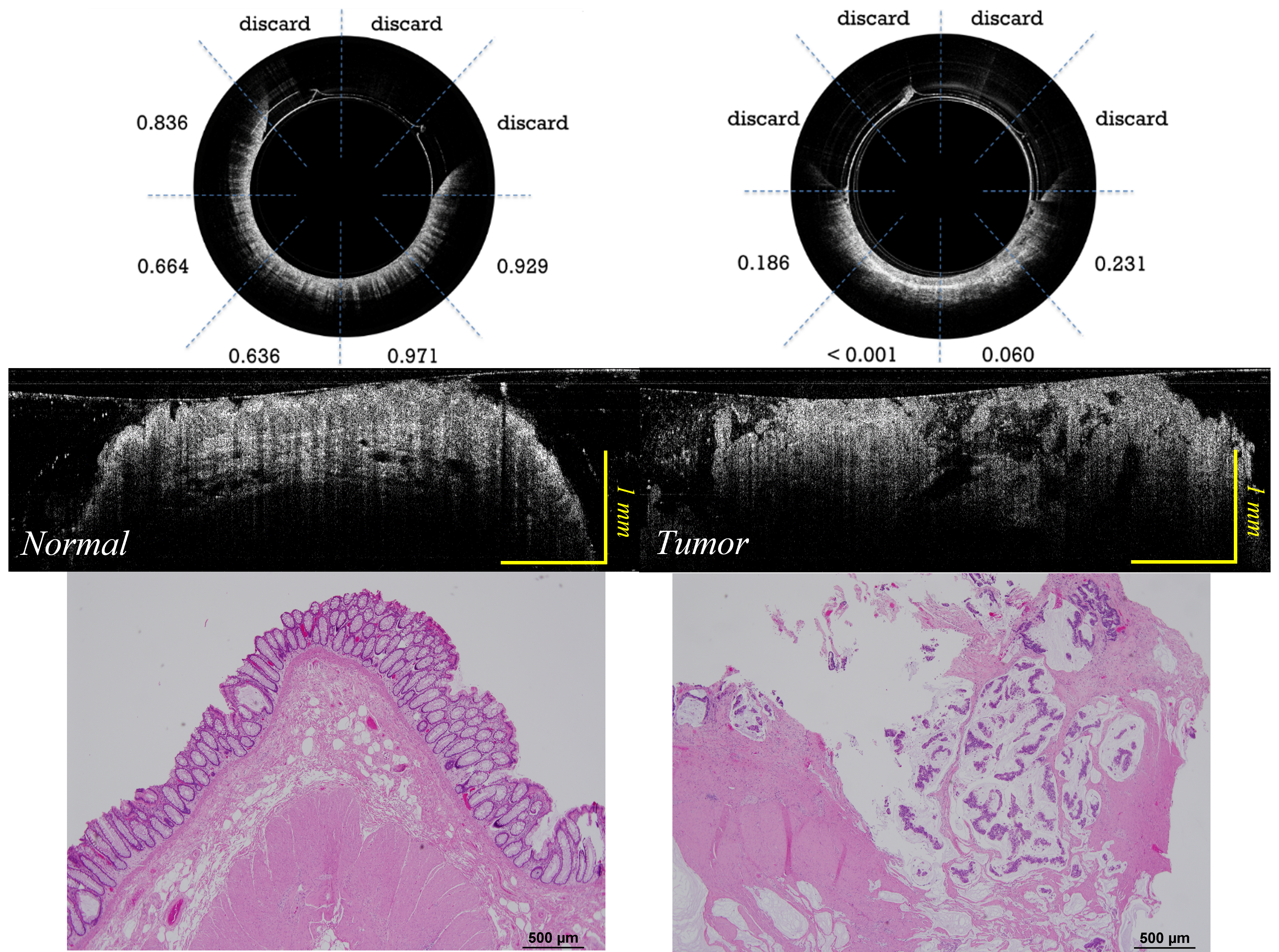
Human coloreactal cancer classification using deep learning and Optical coherence tomography (OCT)
Optical coherence tomography (OCT) can differentiate normal colonic mucosa from neoplasia, potentially offering a new mechanism of endoscopic tissue assessment and biopsy targeting, with a high optical resolution and an imaging depth of ~1 mm. I designed a customized ResNet to classify OCT catheter colorectal images. An area under the receiver operating characteristic (ROC) curve (AUC) of 0.975 is achieved to distinguish between normal and cancerous colorectal tissue images. [Paper] [Code]

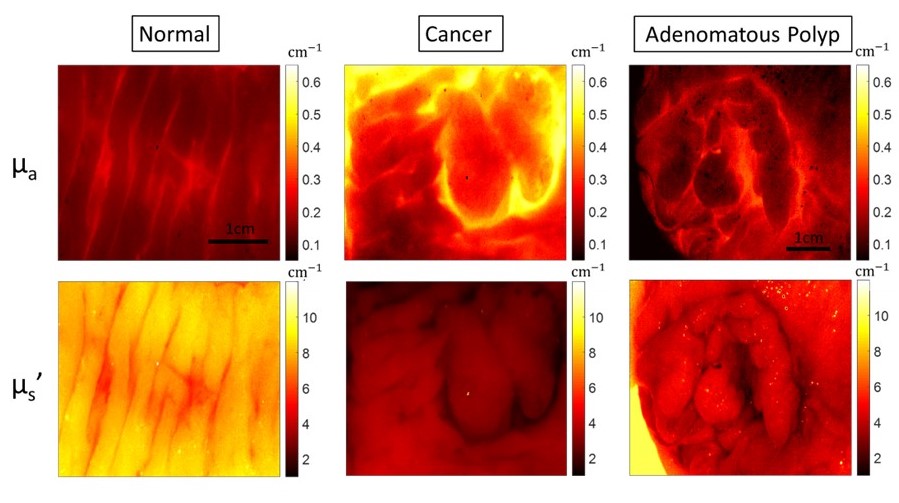
AdaBoost-based multiwavelength Spatial Frequency Domain Imaging (SFDI) for human colorectal tissue assessment
Multiwavelength spatial frequency domain imaging (SFDI) can provide tissue optical absorption and scattering signatures, and would be able to differentiate various colorectal neoplasia from normal tissue. In this ex vivo study of human colorectal tissues, we trained an abnormal vs. normal adaptive boosting (AdaBoost) classifier to dichotomize tissue based on SFDI imaging characteristics. [Paper]
AI-Based Image Reconstruction and Quality Enhancement
Optical imaging like Diffuse Optical Tomography (DOT) offers label-free, radiation-free, and cost-effective solutions for biomedical applications, but its clinical translation has been slowed by long acquisition times, lengthy reconstruction, and inconsistent image quality. To address these limitations, I developed AI-based methods that enhance both image acquisition and reconstruction. I built deep learning models, including physics-informed neural networks (PINNs), that accelerate data acquisition, suppress image artifacts, and improve reconstruction speed and accuracy. Collectively, these advances make optical imaging more robust, efficient, and suitable for clinical use.
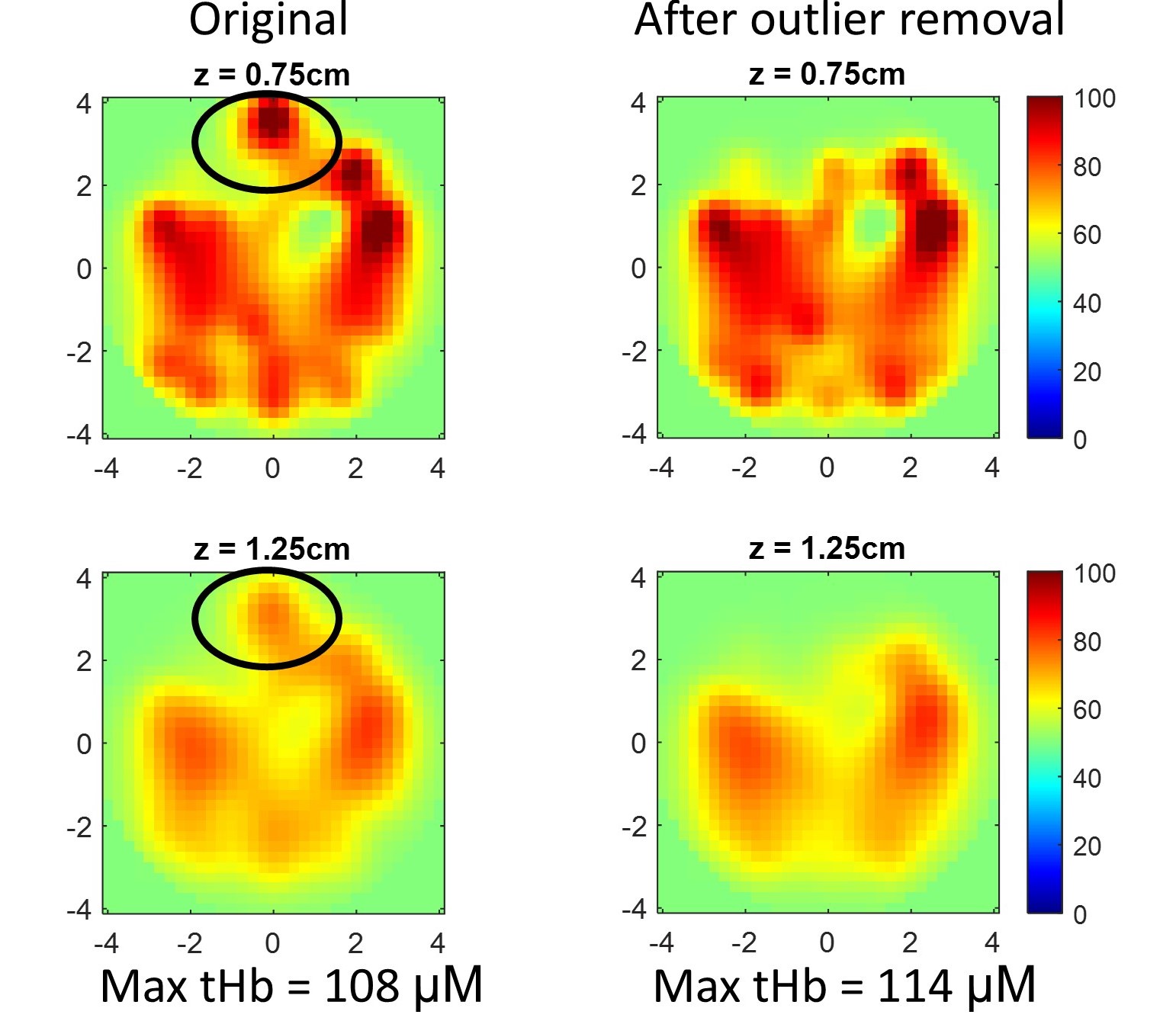
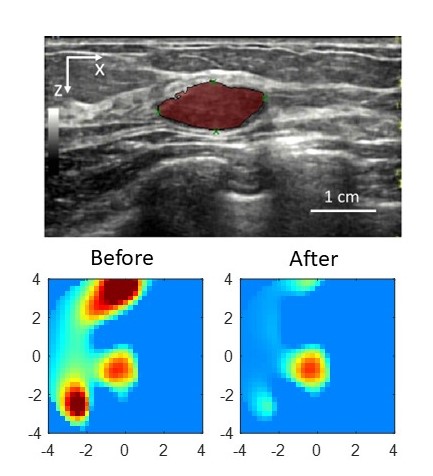
[Paper1:Machine learning model with physical constraints for image reconstruction]
[Paper2: Correction of optode coupling errors]
